This is a post written jointly by Nelson Lau from Brandeis and me, Björn Brembs. In contrast to Nelson’s guest post, which focused on the open data aspect of our collaboration, this one describes the science behind our paper and a second one by Nelson, which just appeared in PLoS Genetics.
![]() Laboratories around the world are generating a tsunami of deep-sequencing data from nearly every organism, past and present. These sequencing data range from entire genomes to segments of chromatin to RNA transcripts. To explore this ocean of “BIG DATA”, one has to navigate through portals of the National Computational Biotechnology Institute’s (NCBI’s) two signature repositories, the Sequencing Read Archive (SRA) and the Gene Expression Omnibus (GEO). With the right bioinformatics tools, scientists can explore and discover freely-available data that can lead to valuable new biological insights.
Laboratories around the world are generating a tsunami of deep-sequencing data from nearly every organism, past and present. These sequencing data range from entire genomes to segments of chromatin to RNA transcripts. To explore this ocean of “BIG DATA”, one has to navigate through portals of the National Computational Biotechnology Institute’s (NCBI’s) two signature repositories, the Sequencing Read Archive (SRA) and the Gene Expression Omnibus (GEO). With the right bioinformatics tools, scientists can explore and discover freely-available data that can lead to valuable new biological insights.
Nelson Lau’s lab in the Department of Biology at Brandeis has recently completed two such successful voyages into the realm of genomics data mining, with studies published in the Open Access journals of Nucleic Acids Research (NAR) and the Public Library of Science Genetics (PLoSGen). Publication of both these two studies was supported by the Brandeis University LTS Open Access Fund for Scholarly Communications.
In this scientific journey, we made use of important collaborations with labs from across the globe. The NAR study used openly shared genomics data from the United Kingdom (Casey Bergman’s lab) and Germany (Björn Brembs’ lab). The PlosGen study relied on contributions from Austria (Daniel Gerlach), Australia (Benjamin Kile’s lab), Nebraska (Mayumi Naramura’s lab), and next door neighbors (Bonnie Berger’s lab at MIT).
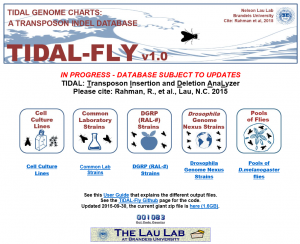
In the NAR study, Lau lab postdoctoral fellow Reazur Rahman and the Lau team developed a program called TIDAL (Transposon Insertion and Depletion AnaLyzer) that scoured over 360 fly genome sequences publicly accessible in the SRA portal. We discovered that transposons, also known as jumping genetic parasites, formed different genome patterns in every fly strain. There are many thousands of transposons throughout the fly genome. The vast majority of these transposons share a virus origin, being retrotransposons. Even though most of these transposons are located in the intergenic and heterochromatic regions of the fly genome, with on average more than two transposons per fly gene, it is a straightforward assumption that some of them are bound to influence gene expression in one way or another.
We discovered that common fly strains with the same name but living in different laboratories turn out to have very different patterns of transposons. This is surprising because many geneticists have assumed that the so-called Canton-S or Oregon-R strains are all similar and thus used as a common wild-type reference. In particular, we were able to differentiate two strains which had only been separated very recently from each other, indicating rapid evolution of these transposon landscapes.
Our results lend some mechanistic insight to behavioral data from the Brembs lab which had shown that these sub-strains of the standard Canton-S reference stock can behave very differently in some experiments. We hypothesize that these differences in transposon landscapes and the behavioral differences may reflect unanticipated changes in fly stocks, which are typically assumed to remain stable under laboratory culture conditions. If even recently separated fly stocks can be differentiated both on the genetic and on the behavioral level, perhaps this is an indication that we are beginning to discover mechanisms rendering animals much more dynamic and malleable than we usually give them credit for. Such insights should not only convince geneticists to think twice and be extra careful with their common reference stocks, it may also provide food for thought for evolutionary biologists. In addition, we hope to utilize the TIDAL tool to study how expanding transposon patterns might alter genomes in aging fly brains, which may then explain human brain changes during aging.
Screenshot of the TIDAL-Fly website:
Given the number of potentially harmful mobile genetic elements in a genome, it is not surprising that counter-measures have evolved to limit the detrimental effect of these transposons. So-called Piwi-interacting RNAs (piRNA) are a class of highly conserved, small, noncoding RNAs associated with repressing transposon gene expression, in particular in the germline. In the PLoSGen study, visiting scientist Gung-wei Chirn and the Lau lab developed a program that discovered expression patterns of piRNA genes in a group of mammalian datasets extracted from the GEO portal. Coupling these datasets with other small RNA datasets created in the Lau lab, the team discovered a remarkable diversity of these RNA loci for each species, suggesting a high rate of diversification of piRNA expression over time. The rate of diversification in piRNA expression patterns appeared to be much faster than in that changes of testis-specific gene expression patterns amongst different animals.
It has been known for a while that there is an ongoing evolutionary arms race between transposon intruders and the anti-transposon police, the piRNAs. In mammals, however, the piRNAs appear to diversify according to two different strategies. Most of the piRNA genomic loci discovered in humans were quite distinct from those in other primates like the macaque monkey or the marmoset and seemed to evolve just as quickly as, e.g. Drosophila piRNA genes. On the other hand, a separate, smaller set of these genomic loci have conserved their piRNA expression patterns, extending across humans, through primates, to rodents, and even to dogs, horses and pigs.
These conserved piRNA expression patterns span nearly 100 million years of evolution, suggesting an important function either in regulating a transposon that is common among most if not all eutherian mammals, or in regulating the expression of another, conserved gene.
To find the answer, the Lau lab studied the target sequences of different conserved piRNAs. One of them was indeed a conserved gene in eutherian mammals, albeit not one of a transposon, but of an endogenous gene. In fact, most of the conserved piRNA genes were depleted of transposon-related sequences. A second approach to test the function of conserved piRNAs was to analyze two existing mouse mutations in two piRNA loci. The results showed that the mutations indeed affected the generation of the piRNAs, and these mice were less fertile because their sperm count was reduced. Future work will explore how infertility diseases may be linked to these specific piRNA loci. It also remains to be understood how a gene family originally evolved as transposon police could evolve into a mechanism regulating endogenous genes.
In summary, this work is an example of how open data enables and facilitates novel insights into fundamental biological processes. In this case, these insights have taught us that genomes are much more dynamic and diverse than we have previously thought, with repercussions not only for the utility any single reference genome can have for research, but also for the role of sequencing individual genomes in personalized medicine.
Rahman R, Chirn GW, Kanodia A, Sytnikova YA, Brembs B, Bergman CM, & Lau NC (2015). Unique transposon landscapes are pervasive across Drosophila melanogaster genomes. Nucleic acids research PMID: 26578579, DOI: 10.1093/nar/gkv1193
Chirn, G., Rahman, R., Sytnikova, Y., Matts, J., Zeng, M., Gerlach, D., Yu, M., Berger, B., Naramura, M., Kile, B., & Lau, N. (2015). Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals PLOS Genetics, 11 (11) DOI: 10.1371/journal.pgen.1005652












Comments are closed.