The FoxP gene family comprises a set of transcription factors that gained fame because of their involvement in the acquisition of speech and language. While early hypotheses circulated about its function as a ‘learning gene’, a simultaneous “motor-hypothesis” stipulated that the gene may be more of a motor learning gene, involved in different kinds of motor learning, one of which is speech acquisition. Work in animals as diverse as mice and fruit flies over the last 20 years has firmly established at least some of the FoxP genes as crucial for motor learning tasks that are not involved in language.
Now, our graduate student Ottavia Palazzo (with invaluable support and training from Mathias Rass from the neighboring Schneuwly lab) has generated and thoroughly characterized a set of new transgenic fly lines to help us better understand the role of FoxP in the form of motor learning we are studying, operant self-learning.
Using CRISPR/Cas9 with homology-directed repair, she tagged the FoxP gene in two different ways. In one line, she tagged the gene such that we can express a fluorescent protein in all neurons where any of the three different isoforms is expressed, that this gene can give rise to. In the other line, she tagged only the one isoform that we think is associated with operant self-learning.
This one isoform is expressed throughout the adult brain of the fly, but not in the mushroom bodies, where a few previous reports had detected it, using a technique which can sometimes lead to incorrect expression patterns. In fact, because three previous studies reported three different expression patterns for the same gene, we chose this particular tagging strategy to avoid the problems associated with this technique. Ottavia found about 1200 neurons expressing the isoform we were interested in:
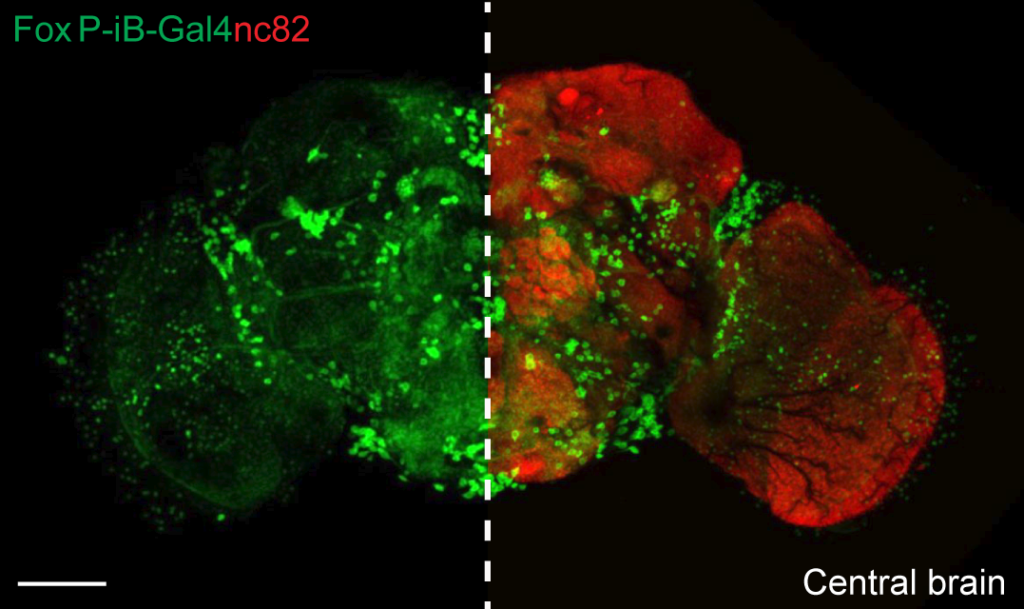
Contrasting the expression of this isoform with the expression of the other two isoforms, revealed an additional ~600 neurons which express one or both of them (marked in blue):
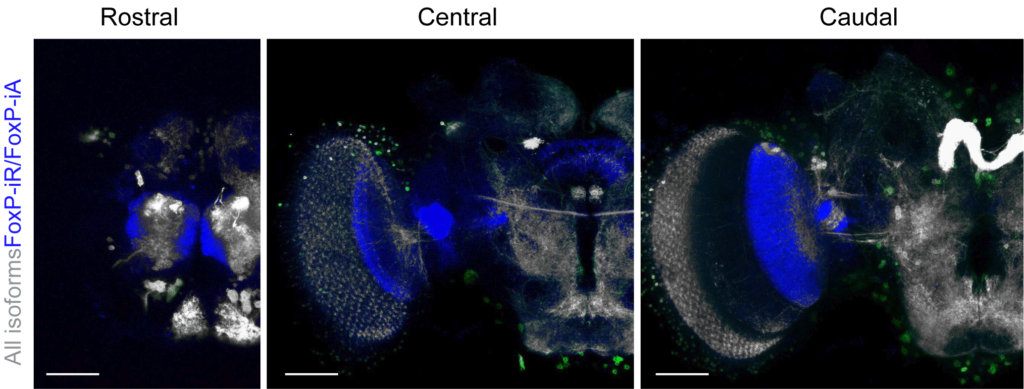
Also here, not a hint of any expression in the mushroom bodies. Given the previous reports, we looked particularly closely not only in adults, but also in larvae, but could not find any expression. We also used an antibody (which we verified against mutants and our tagged lines to be highly specific) and found no expression in mushroom bodies. Our results with two different genomically tagged lines and the antibody corroborate earlier work with a differently tagged line and a reporter gene approach, which also failed to detect expression in the mushroom bodies. Given the multitude of different approaches all converging on identical expression patterns, it seems now clear that mushroom body Kenyon cells do not express FoxP above the detection threshold of these techniques. If the levels of expression that we detect are necessary for the physiological function of FoxP, it is conceivable that any expression below these thresholds may likely be physiologically irrelevant.
To see if there are any general problems with these fly lines (which may be problematic for the subsequent learning experiments), Ottavia tested the flies in Buridan’s paradigm. In case you haven’t heard about this experiment, here’s a short video I made 10 years ago:
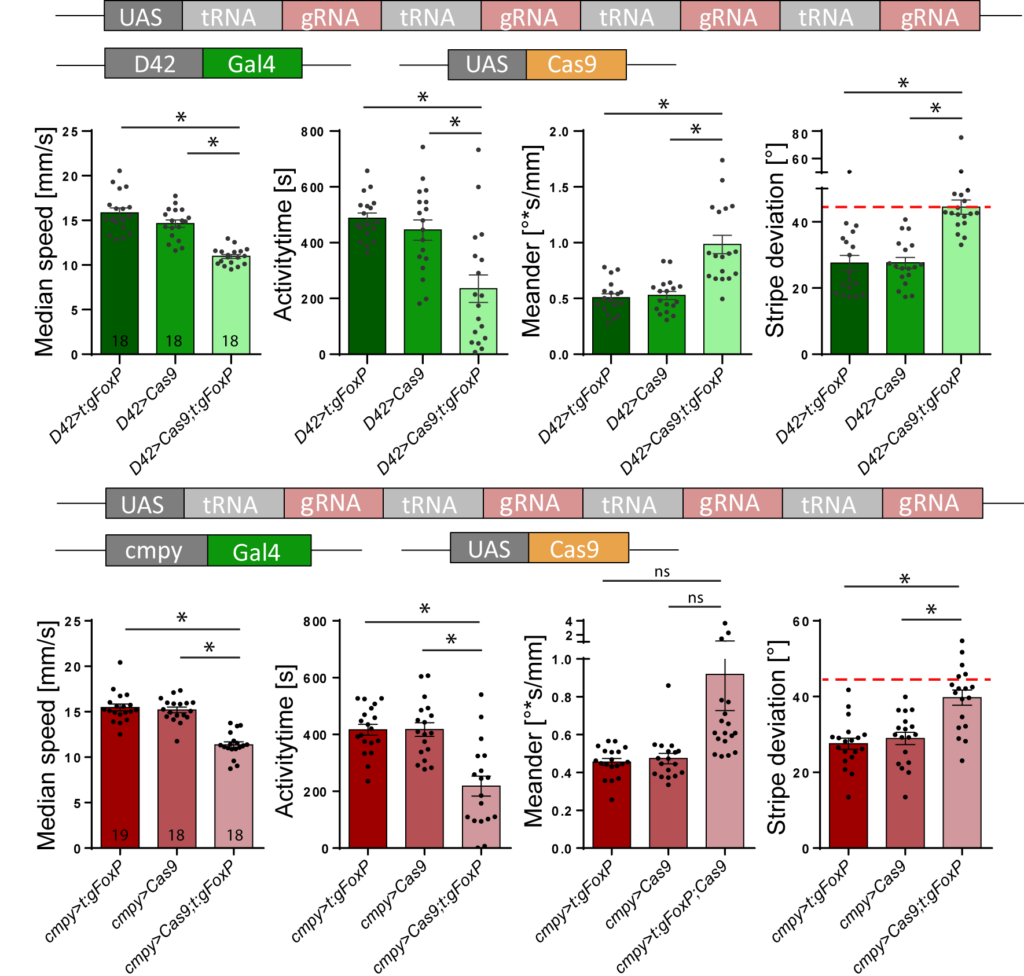
Not unexpectedly, she found that the insertions she had made disrupted FoxP expression and had substantial effects on walking and landmark fixation:
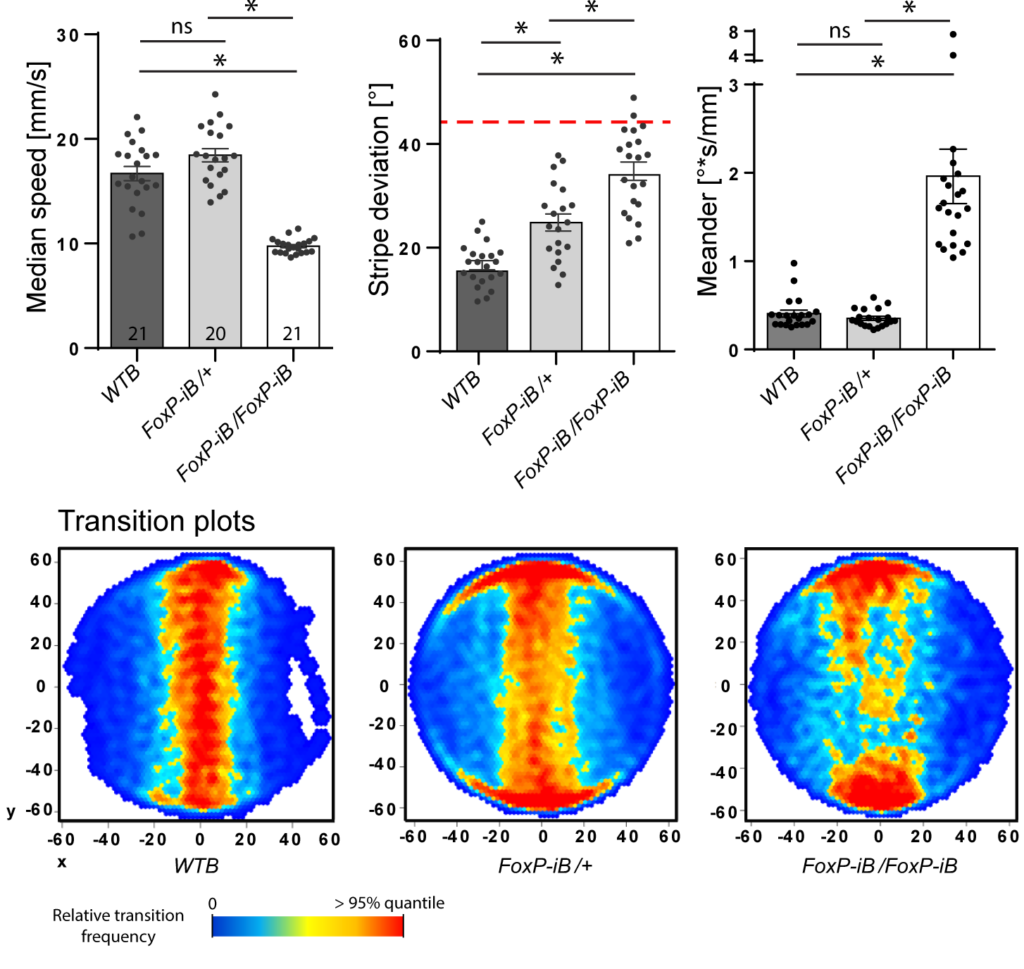
Not only do the flies with homozygous insertions walk more slowly, they also fixate the stripes less and walk less straight (meander).
She also tested one of the original FoxP mutations, the widely used 3955 allele, and found similar defects:
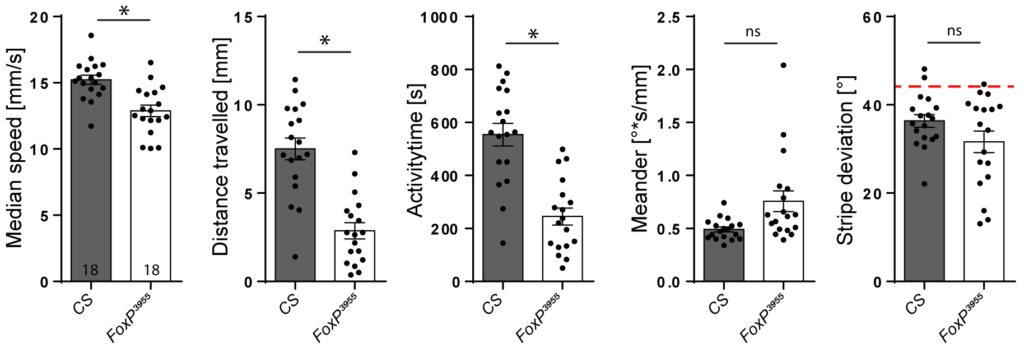
Curiously, one of the studies that had (erroneously?) detected FoxP in the mushroom bodies, failed to detect this rather conspicuous (~20% or more) walking defect in these mutant flies, despite testing for it. At the time, I had already noticed that their control experiments lacked the sophistication to capture the motor defects I thought were most critical for their experiments, but apparently they were not even sensitive enough to detect such major defects. In summary, in this (Science!) paper, the authors detected FoxP where there apparently isn’t any, but failed to detect a severe motor phenotype, despite looking for it.
Ottavia also created a CRISPR line to knock FoxP out when and where she wanted. In one of her experiments, she knocked the gene out in early pupae or adult flies and found that this left walking behavior and landmark fixation of the flies unaffected. In other words, for these behaviors, FoxP is only important during development.
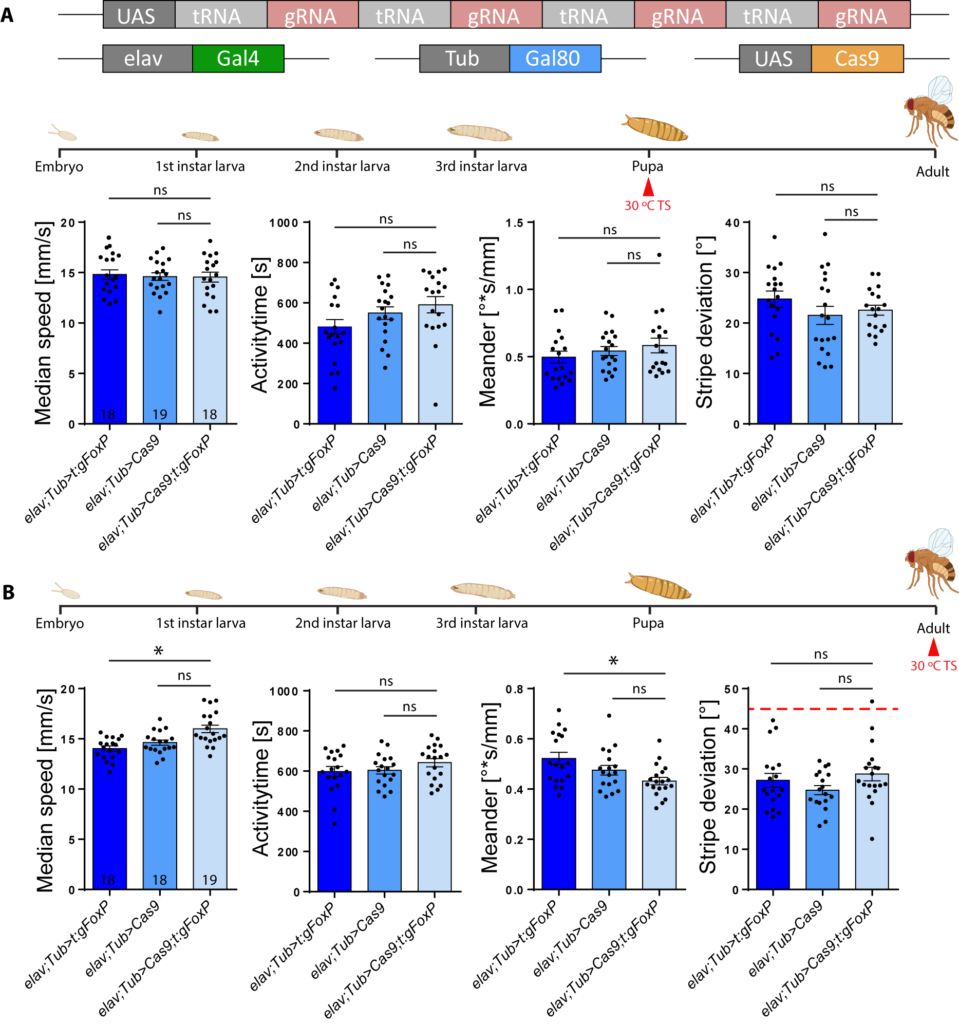
Similarly, knocking FoxP out of dorsal cluster neurons (important for stripe fixation and expressing FoxP) or mushroom body Kenyon cells (important for walking and stripe fixation, but not expressing FoxP), also had no effect:
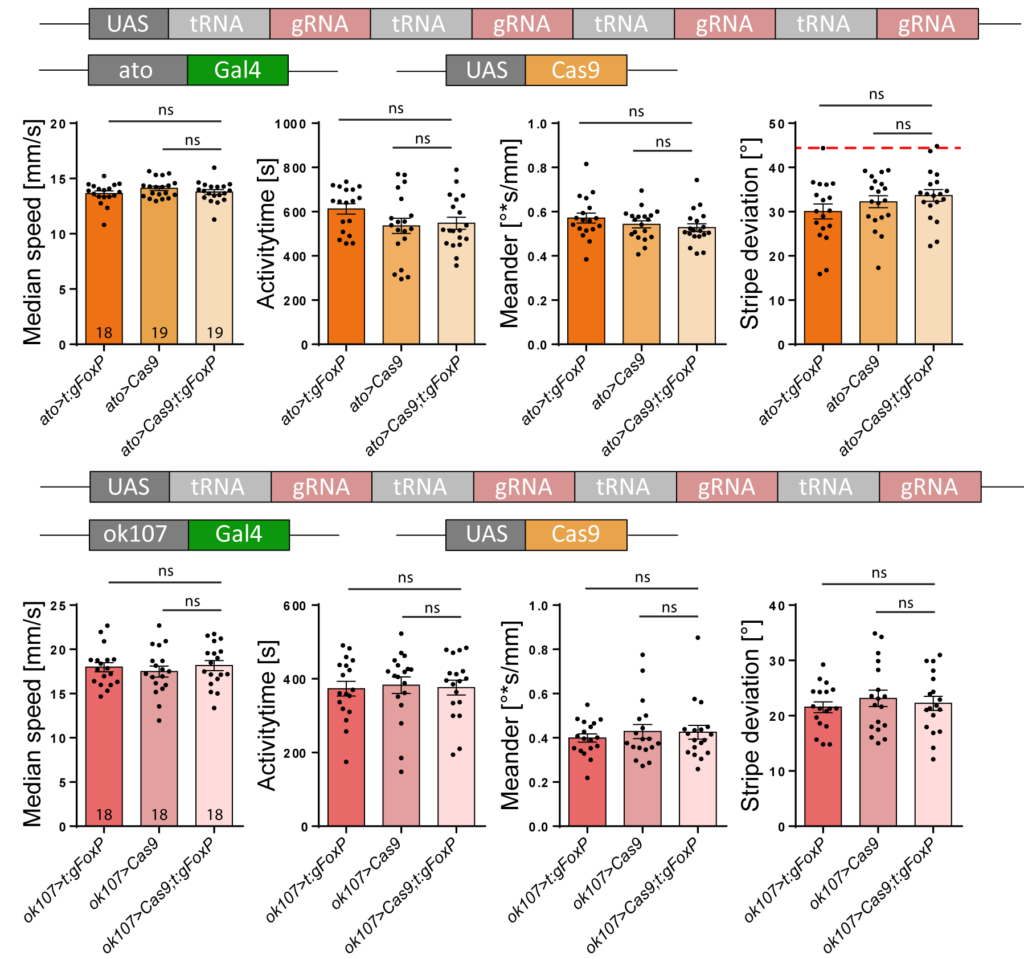
On the other hand, when she excised the FoxP gene from motorneurons (e.g., with the D42 driver) or from neurons in the protocerebral bridge (with the cmpy driver), she saw almost the same effects as if she had deleted the gene constitutively:
The next step in this line of work will be to see which of these manipulations fly enough for the toque learning experiments and to see which neurons need FoxP for operant self-learning.
CRISPR/Cas9 was a new technique for our lab and it worked exceedingly well, both for tagging the gene and for knocking it out. In our hands, it worked with high efficiency, reliably and, as far as we can tell, with no off-target effects.
The results here also contradict some prominent publications in our field, so it will be interesting what, if anything, is going to happen to reconcile the findings.
Of course, as we try to practice open science, all the raw data for this work is publicly accessible with a liberal re-use license.