![]() The data clearly show that publications in Cell, Nature or Science (CNS for short), on average, cannot be distinguished from other publications, be it by methodology, reproducibility or other measures of quality. Even their citation advantage, while statistically significant, is so small that it is practically negligible. Regardless of all the data, individual examples sometimes serve to illuminate the data and drive some facts home. There are more or less extreme examples of CNS publications not meeting the expectations to which they are commonly held. It’s needless to mention all the examples that went through the media, but I would instead cite a paper from my field (where I feel reasonably competent) as an instance of an extreme case of an obviously flawed paper making in into the journal Nature.
The data clearly show that publications in Cell, Nature or Science (CNS for short), on average, cannot be distinguished from other publications, be it by methodology, reproducibility or other measures of quality. Even their citation advantage, while statistically significant, is so small that it is practically negligible. Regardless of all the data, individual examples sometimes serve to illuminate the data and drive some facts home. There are more or less extreme examples of CNS publications not meeting the expectations to which they are commonly held. It’s needless to mention all the examples that went through the media, but I would instead cite a paper from my field (where I feel reasonably competent) as an instance of an extreme case of an obviously flawed paper making in into the journal Nature.
However, one doesn’t even need to look at the extreme cases. Let’s look at a paper in Science Magazine I actually quite like. It’s an interesting topic, a beautiful experimental paradigm and the results in the wildtype flies are truly exciting. Their mutant and transgenic results, if correct, are tantalizing as they run counter to essentially all available literature on the gene in question. I personally know and like the senior author of the paper, Gero Miesenböck, and appreciate all of his work, including this publication. All I’ll try to do here is to make the case, that this paper is an average paper in terms of the level of evidence provided to support the claims made by the authors. Not obviously worse than most papers in our field, but definitely nowhere near the level of evidence those colleagues ignorant of the data would assume to be required for a CNS paper. All papers have strengths and weaknesses, some have more uncertainty associated with their results, others less. The data suggest that there isn’t much difference between journals, on average, and I’ll try to use some details in this Science paper to exemplify the data.
The authors state in their title that the gene dFoxP influences the speed of perceptual decision-making in Drosophila fruit flies. They test the flies by first conditioning them to avoid a given odor using electric shocks. The choice test is then done in a chamber on individual flies. Two odors enter the chamber from opposite ends and exit the chamber in the middle. The fly then walks back and forth between the ends of the chamber and avoidance is measured by where the fly spends most of the time: in the half with the odor associated with electric shock, or in the half with the control odor. In the current study, the authors made the control odor increasingly similar to the one associated with shock and found that wildtype flies spent more time in the middle of the chamber, where they had to decide of whether to keep walking or make a turn, the more similar the odors were. That is definitely an exciting finding, suggesting that more similar odors require more processing time for an avoidance decision than less similar odors.
In order to get a handle on the biological processes underlying this interesting phenomenon, the authors have conducted a screen of genetically modified flies, looking for mutants with longer decision times than the wildtype flies. One of the candidates that showed up in the screen, is the Drosophila orthologue of a gene involved in language in humans, FoxP. It seems hard to understand what a gene generally thought to be involved in motor learning has to do with this perceptual task, but data are data, unless there are alternative explanations. There are three main concerns that raise the suspicion that the involvement of dFoxP in this task may not be as straightforward as suggested in the title of this paper.
- It s common practice to homogenize the genetic background of mutants with that of the control strain to which the mutants are compared. This is done to exclude variations other than in the gene of interest to be responsible for any differences. However, the authors do not state whether and if so for how many generations the mutant flies were outcrossed to the proper wildtype genetic background. Given that the flies came from a screen, it is unlikely that all strains tested in the screen were outcrossed. For instance, the alleles described in the paper show significant lethality which disappears when outcrossed for six generations, suggesting that their general level of fitness and health appears to be affected by their genetic background. It is thus possible that factors in the genetic background and not dFoxP may be responsible for the phenotype. Given the track record of the Miesenböck lab, it is unlikely they did not outcross the flies, but at the very least, the number of generations for which the outcrossing has taken place would have to be listed in the paper.
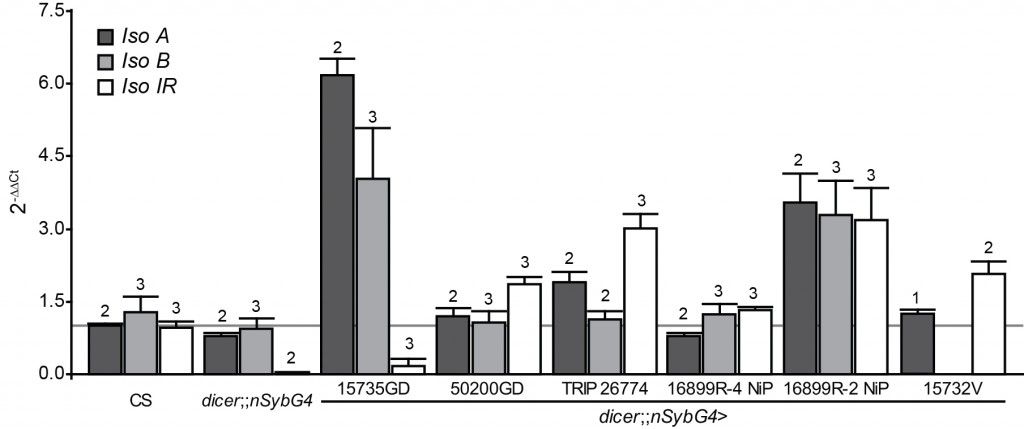
- A common procedure to test for genetic background effects, in addition to outcrossing which can reduce but not eliminate genetic background effects, is to use an additional method to manipulate the gene in question. The authors used RNA interference (RNAi) to attempt to down-regulate the expression of dFoxP. It is common practice to use quantitative PCR to test the effectiveness of this method, as a) RNAi usually does not abolish gene expression completely and b) it is known to have off-target effects, i.e., it may affect the expression of other genes besides/instead of the target gene. The line used in this study comes from a collection known to have problems with off-target effects, so testing effectiveness is especially important. However, no evidence that such a test was performed is described. Below is what such data would look like, if you were to order all RNAi constructs currently available, drive their expression in all neurons (with nSyb-GAL4) and then have an undergraduate student run a whole slew of qPCR reactions to compare the expression (in fly heads) of all three dFoxP isoforms to the respective control strain (many thanks Joel and postdoc Axel for all that work!): No effect!
I cannot emphasize enough that the graph above represents very preliminary data. It’s data from one undergraduate student who had prior experience with qPCR before coming to our lab, but who did not do this work full-time. There definitely need to be more biological replicates and some additional drivers need to be used (e.g. an actin driver to drive RNAi in all cells). On the other hand, the results match my own qPCR results with these lines and results from another laboratory that we collaborate with. So while I would not exclude that we may see significant reduction in dFoxP expression with some technical tweak, the currently available data suggest that none of the available dFoxP RNAi lines significantly knocks down dFoxP expression, including the one tested by DasGupta et al. (15732V). Our work also points to a reason why none of the lines seems to have a detrimental effect on dFoxP expression: we have localized polymorphisms in the dFoxP gene which could bias the RNAi process towards sequestration, rather than degradation of the mRNA, potentially explaining some of the very high values in the plot above. Thus, it is possible (and we consider it even very likely) that some or all of these lines actually do affect dFoxP expression in the way intended. However, we currently have little possibility to ensure it’s not an off-target effect after all.
I’d thus tentatively conclude that the phenotype DasGupta et al. have discovered can indeed be ascribed to dFoxP action in this task. However, at this point, there is insufficient data to make that statement with any reasonable certainty (no matter how likely I personally find that to be the case!). - Finally, previous work on FoxP both in other animals and in flies (including our own paper coming out tomorrow, watch this space after 5pm EST on June 25) suggests an involvement of dFoxP in motor learning and perhaps also (consequently?) motor coordination. In fact, our work shows that learning about external stimuli is fine in FoxP mutant flies. It is thus critical to make sure that any animals showing a deficit in this task can perform the required movements (walking, turning, starting, stopping) accurately. The authors here attempted to ensure this by measuring the time spent in sections of the chamber other than the decision section in the middle. However, one would assume that flies will mostly turn or sit at the ends of the chamber and in the decision sector, with fewer turns and pauses in the sectors in-between. Thus, by evaluating sectors in which the flies mainly walk straight, the authors may be underestimating the contribution of turns and no-movement episodes to the decision-making process. If that is correct (and the brief explanations in the methods section make it very difficult to be entirely sure about how precisely and on which sectors they have performed the calculations), dFoxP-manipulated flies may either have a problem starting to walk from a stop, or have difficulty turning in the chamber and the authors would interpret this as an increased decision-time, rather than a motor problem.
In summary, it may well be that dFoxP entails all the functions reported by DasGupta et al. and this would indeed make for a giant head-scratcher about the (ancestral) functions of FoxP, given all the mountain of work involving it in motor, rather than perceptual processes. However, at this point, some more work (at least some more detailed method explanations!) is required to be reasonably certain about the existence of such a problem. Personally, I do think that the phenotype is due to the authors’ FoxP manipulations, but I may be wrong, the evidence is not strong enough either way. I do have my doubts, however, about the phenotype not being due to motor defects, but I could be wrong there as well, as the data is inconclusive. At this point, we need more experiments and/or data analysis to show who is wrong. Again, I still like the paper, it’s the kind of work that drives our field forward.
Speaking more generally: there is no need for any researcher to wait (perhaps indefinitely?) until they have the perfect data set, with the unambiguous results and the foolproof conclusions. Just send your plain decent work to CNS magazines as well! If they publish it, you will get a job and nobody but some lone blogger will ever ask about the content of that paper ever again.
DasGupta, S., Ferreira, C., & Miesenbock, G. (2014). FoxP influences the speed and accuracy of a perceptual decision in Drosophila Science, 344 (6186), 901-904 DOI: 10.1126/science.1252114













Dr. Brembs,
Thank you for that insight. Finding the most suitable journal may include a methodology of using more than two journals to discern the leading choice journal for one’s given research for review and publication. Nonetheless, any number of journals in a given topic cluster should be considered for potential review and publication.
Jonah Lissner
Independent Researcher of Science, PhD Student
https://lissnerresearch.weebly.com
People really need to realize: Science/Nature are for ‘exciting’ results, Neuron (and I had thought Cell) are for in-depth, less sexy results.
well, there is more space in these journals, but basically Glam nonetheless.
So if Science publishes my paper (lets say studying operant learning in snails), i can get a good job and start a blog ranting about other people trying the same? Only to ensure i got it correctly. However i agree with most of the posts on this blog. Keep up the good work!
Ah, I think double@standards from Universität Köln misunderstood the main thrust of the post: I don’t want to discourage people to try the same, I want to encourage them: send ALL your work to GlamMagz! You are dead on, it worked for me: our Science paper on operant conditioning in snails was a paper with ZERO scientific novelty: it only confirmed what we already knew. And now I have a job and try to get rid of this silly way of assigning cred.
Thus, quite the opposite: I’m not ranting about other people, I’m showing an example of what the data show: GlamMagz are unscientific and any means to bring down this system is valid. I wonder what would happen if we really all sent all our papers to GlamMagz first. I almost think we should do that, if nothing else works.
Totally agree, always been a fan of your cause. Also always been a fan of asking critical and uncomfortable questions. Your reply does not fail to satisfy! Keep up the good work and say hi to your spouse (who i will probably meet in the US in 2 weeks if i read the abstract booklet correctly). All the best, Alex A. 🙂