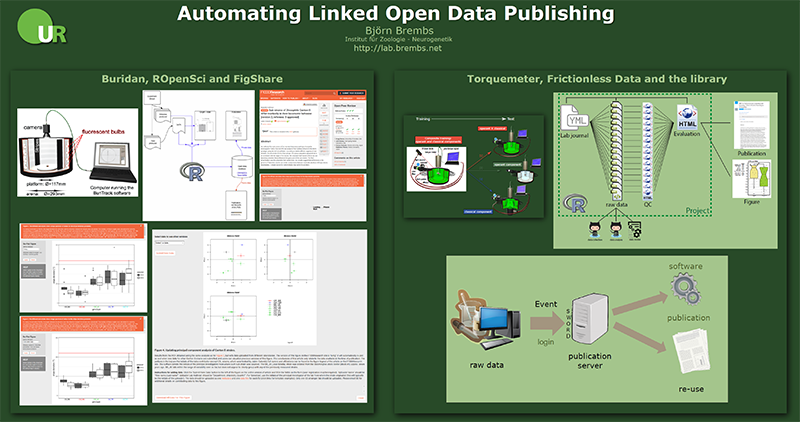
On the occasion of the first “BigDataDay” at our university, I have summarized on the below poster our two main efforts to automate the publication of our tiny raw data.
On the left is our project automating Buridan data deposition at FigShare using the ROpenSci plugin and the consequence of just sending the links to the data and the evaluation code to a publisher, instead of pixel-based figures, when submitting a paper. Most of this work was done by postdoc Julien Colomb several years ago, when I was still in Berlin.
On the right is our ongoing project of automating flight-simulator data deposition with our library. We designed a YML/XML meta-data format loosely based on the Frictionless Data standard. Our evaluation script reads a YAML file that contains the experimental design (i.e., which raw data file belongs into which experimental group) as well as formalized commands for the kinds of statistical tests and graphs to be generated. From this information, each experiment (i.e., XML file) is evaluated and a quality control HTML document is written that contains numerous aspects of the raw data to ensure the quality of the data in each experiment. The same information from the YAML file is used to compute an evaluation HTML document with group-wise evaluations. All the raw data and evaluation files are linked with each other and the XML files link not only to the repository with the evaluation script, but also to the repository with the software that collected the data and the data model explaining the variables in the raw data files. Ideally by dragging and dropping figures with statistics into a manuscript, published scholarly articles would link to all of the files generated for the publication. A client-side Python script is called upon user login to compare the local project folder and compare it with the folder on the library’s publication server for synchronization.